The Association Between Interleukin-7 and Immune-Related Adverse Effects in Immune Checkpoint Blockade Cancer Treatment
Immune checkpoint blockades (ICBs) have become an emerging cancer therapy. Many may have heard of the unexpected yet outstanding clinical trial where every colorectal cancer patient (n=18) experienced full remission after ICB treatment. Even so, ICB is not a universal cure for cancer–its efficacy is not guaranteed in every individual, and it may lead to severely harmful effects. These two papers, released within days of each other, aimed to determine certain genetic markers that may predispose someone to severe responses to ICB, known as immune-related adverse effects.
First, it’s important to understand the source behind ICBs' volatile power. As the name suggests, ICBs suppress the regulatory checkpoints in our immune system in turn releasing the “break” on our immune response. How does it do this? ICB is a monoclonal antibody, a protein that marks foreign antigens as hazardous. During cancer, tumor cells are often bound to neutralizing antibodies allowing them to escape the surveillance of the immune police. However, when someone is treated with ICB, the monoclonal antibodies are able to bind to tumor cell receptors, blocking the binding site of neutralizing molecules, and ultimately signaling T cells to come to destroy the tumor cells.
When this treatment works, it is spectacular, leading to full remission and recovery, as seen in the colorectal cancer patients. However, with threshold inhibition raised higher, this poses the risk of autoimmunity and toxicity. Why can’t these antibodies bind to normal cells and mark them for destruction? Why doesn’t the immune system start overreacting due to its increased sensitivity? When this does happen, it is known as immune-related adverse effects (irAEs), and researchers turned to genetics for an answer.
From a cohort of 1,751 European patients, researchers employ a Genome-Wide Association study (GWAS) to identify certain genetic variations, known as single nucleotide polymorphisms (SNPs), correlated with either severe or all-grade irAEs. Though a significant SNP for severe irAES was not identified, GWAS revealed 3 SNPs associated with all-grade irAE, the top result within an intron (a non-coding region of the DNA) in the interleukin-7 (IL-7) gene. This gene encodes for the interleukin-7 cytokine, a protein that signals an immune response. Interestingly, IL-7 has been known to aid in T-cell proliferation, differentiation, and function as well as contribute to pro-immune response. Though this may seem beneficial at first, the function of IL-7 may lead to the overreaction of the immune system, known as a cytokine storm, and may lead to an explanation of the root cause of irAEs.
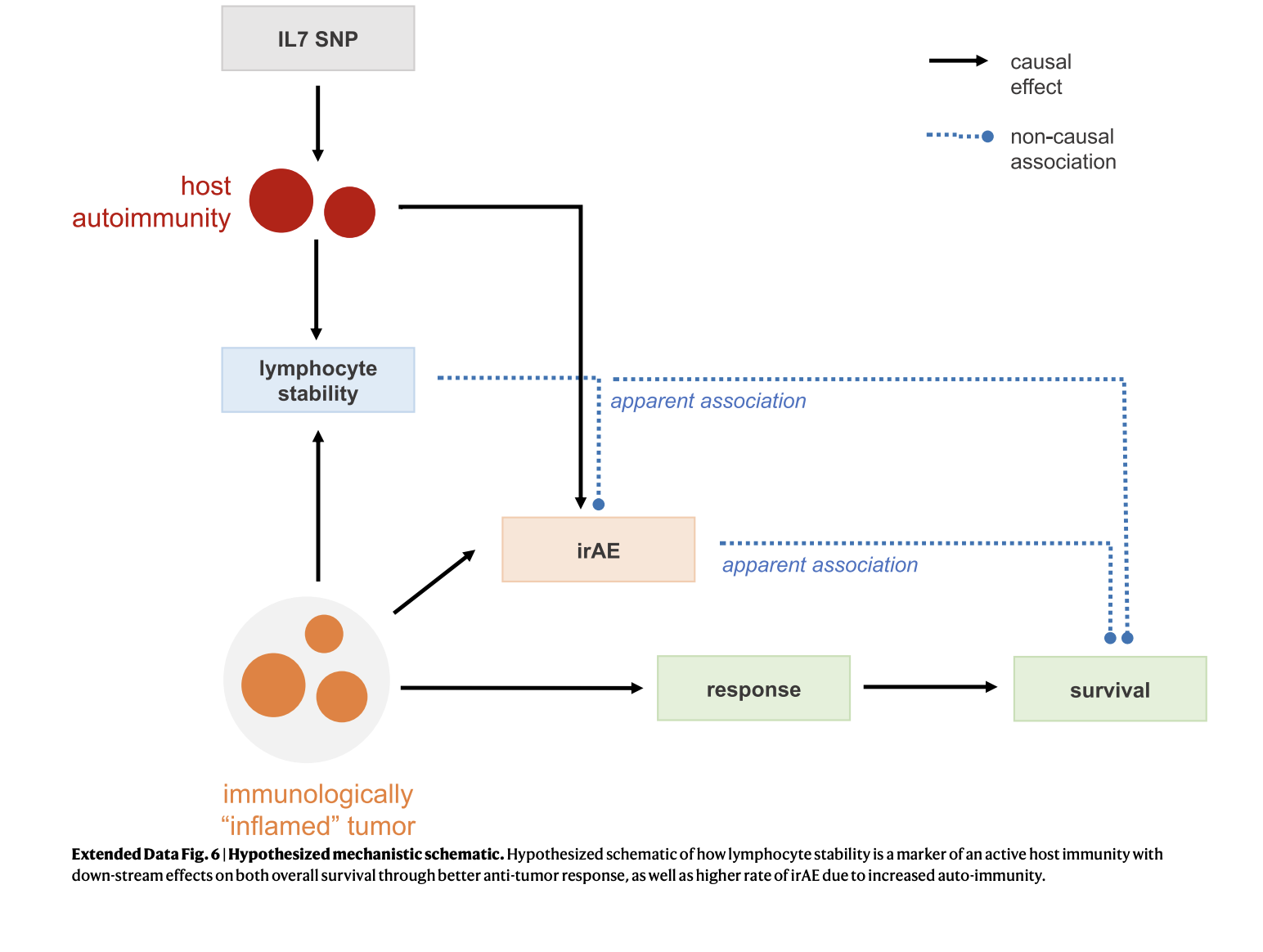
After logically providing an explanation of the association between irAEs and an SNP in the IL-7 gene as well as continuing to verify this through clinical validation across hospitals and adjusting for confounding factors such as sex, age, and pre existing disease, researchers aimed to find the underlying mechanism of how the change in one nucleotide base pair amidst the entire genome, could determine how someone would respond to cancer treatment.
Ultimately, researchers identified a novel cryptic exon as a putative mechanism for the SNP. The human genome consists of coding exons and noncoding intron; however, certain genes may be expressed during the changes that occur due to alternative splicing. Having a slight variation in this particular base pair leads to alternative splicing, resulting in the synthesis of a 70-base pair cryptic exon (IL7CE). This extra exon, formed as a result of having the SNP, is highly expressed in B-cells and moderately expressed in T-cells. Furthermore, it is correlated with total IL7 expression, the coexpression of IL7:IL7R, and suggests that IL7 expression stabilizes lymphocyte counts and enhances lymphocyte homeostasis. Many of these factors are positive features, in moderation. In fact, the study found that if an individual obtains the SNP but does not contract irAEs, they obtain higher survival rates than individuals who do not have the SNP. However, due to the extra exon expressed in B cells in turn controlling T-cell activity, individuals with the IL-7 SNP face greater susceptibility to irAEs.
The second paper delved even deeper into the mechanisms behind the SNP. IL-7 expression was correlated with B cell transcription factors, including IKZF3 and POU2AF1, in both healthy donors and untreated patients and consequently drove enhanced activation and maturation of B cells. In fact, pathway analysis was enriched for B-cell maturation pathways inducing “immunoglobulin prosecution” and “positive regulation” of B-cell activation. On the other hand, researchers found that the anticorrelated genes were associated with TLR4 signaling, neutrophil degranulation, and antigen presentation.
Secondly, since ICB treatment typically affects T-cell function, researchers aimed to determine how and if IL-7 obtained an effect on them. Since B-cells are important activators of T cells and IL-7 in B-cells associate with T-cell responses, this offers the first piece of information to understand the mechanism. The study revealed that after ICB treatment, carriers of the SNP exhibited increased terminally differentiated effector memory (TEMRA) T cells that then re-express CD45RA, a gene associated with differentiation T cells, and a decrease in naive T cells. After quantification, researchers found an increase in intermediate differentiated T cells, whose function typically involves the “killing” of foreign cells, in individuals with the SNP compared to those without the SNP. These findings seek to explain the intricate mechanistic details by describing how the presence of the SNP affects B-cell maturation and activity which leads to T-cell populations’ ability to kill cancer cells.
Finally, and perhaps most importantly, researchers found the SNP to be associated with post-immune checkpoint blockade CD8+ T-cell clonality, a kind of readout for immune response. This makes sense as the SNP leads to a stable lymphocyte population and high lymphocyte stability leads to more large clones. This was shown in their study which examined CD8+ T-cell clone sizes, based on the unique TCR CDR3 nucleotide sequences. They also employed Gini coefficient, a measure of inequality, and found SNP carriers showed heightened levels of this uneven distribution of T-cells.
Even with this plethora of fascinating data, I am still curious about the structural components, such as information shape and size, which could potentially be accomplished through a Western blot, or information on chromatin availability in the region to truly solidify our understanding of this mechanism. Regardless, this research has offered truly profound and quality data and contributions to the fields of research and medicine. Under the bright yet distant light of personalized medicine, identifying and explaining why and how microscopic variations in the genetic sequence function may lead to the minimization of potentially fatal side effects and foster greater efficiency in treatments. And perhaps, we may be able to reach a day where the clinical trial with 100% remission rates, or even eradication, may be a reality for everyone.
References
Groha, S., Alaiwi, S.A., Xu, W. et al. Germline variants associated with toxicity to immune checkpoint blockade. Nat Med 28, 2584–2591 (2022). https://doi.org/10.1038/s41591-022-02094-6
Taylor, C.A., Watson, R.A., Tong, O. et al. “IL7 genetic variation and toxicity to immune checkpoint blockade in patients with melanoma.” Nat Med 28, 2592–2600 (2022). https://doi.org/10.1038/s41591-022-02095-5