Have a BLAST with Sequence ALignment Part II
With the click of a few buttons, you can now perform BLAST yourself through the NCBI website: https://blast.ncbi.nlm.nih.gov/Blast.cgi. Below is a step-by-step guide on how to navigate through the website to successfully BLAST off!
What you will need:
A computer
The nucleotide sequence from your favorite gene or protein
Open the website and select “Nucleotide BLAST” for DNA sequences of “Protein BLAST” for protein sequences.
Copy and Paste your sequence (you can find many in the NCBI Gene Bank) into the first box.
If you only want to compare a certain number of base pairs, you can delineate this by filling in the “Query Subrange”
Add a title into the “Job Title” box.
Report what organism the gene or protein sequence came from using its scientific name (i.e. “Homo sapiens” not “Humans”).
Exclude low-quality samples or unvalidated sources by selecting the checkboxes next to “Exclude” and/or “Limit” (optional)
Select the quality of BLAST to run: megablast, discontiguous megablast, or blastn
For additional edits, click the dropdown on “Algorithm Parameters.” Here, you can do anything from adjusting the word size to decreasing mismatches by changing the numbers in mismatch scores.
Countdown, take a deep breath, and brace yourself for the immense power and wonders of BLAST. Once you feel ready, slide your mouse on top of the vibrant blue BLAST button and press down to BLAST off.
You can see the most similar sequence identified by BLAST by clicking the first link and pairwise alignments should appear.
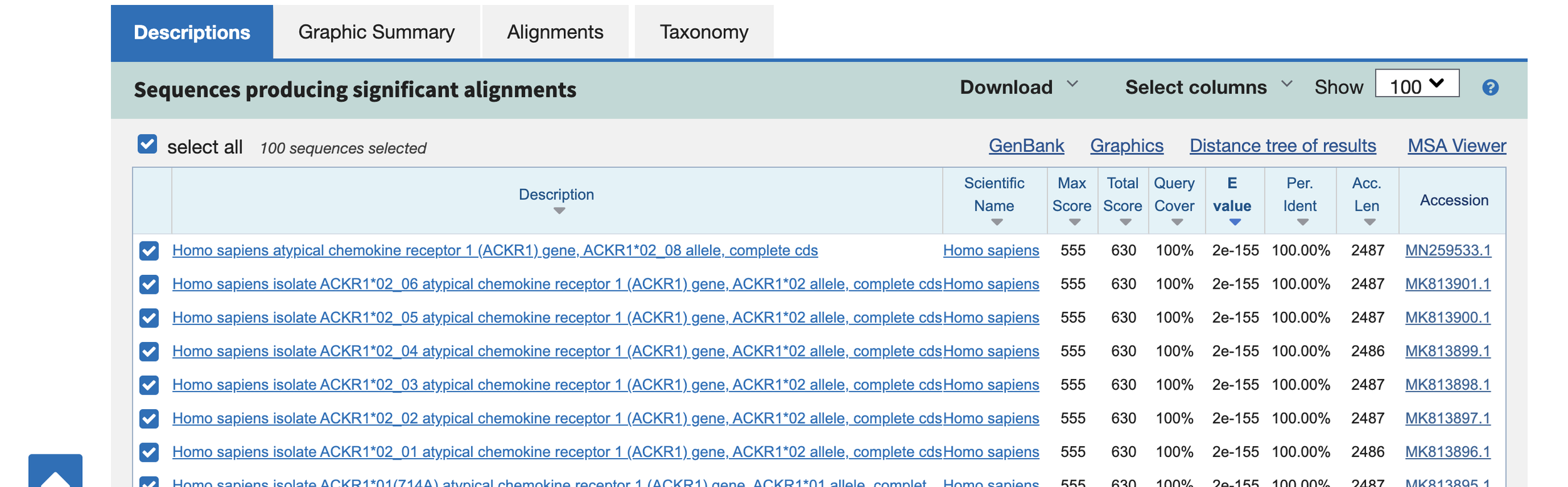
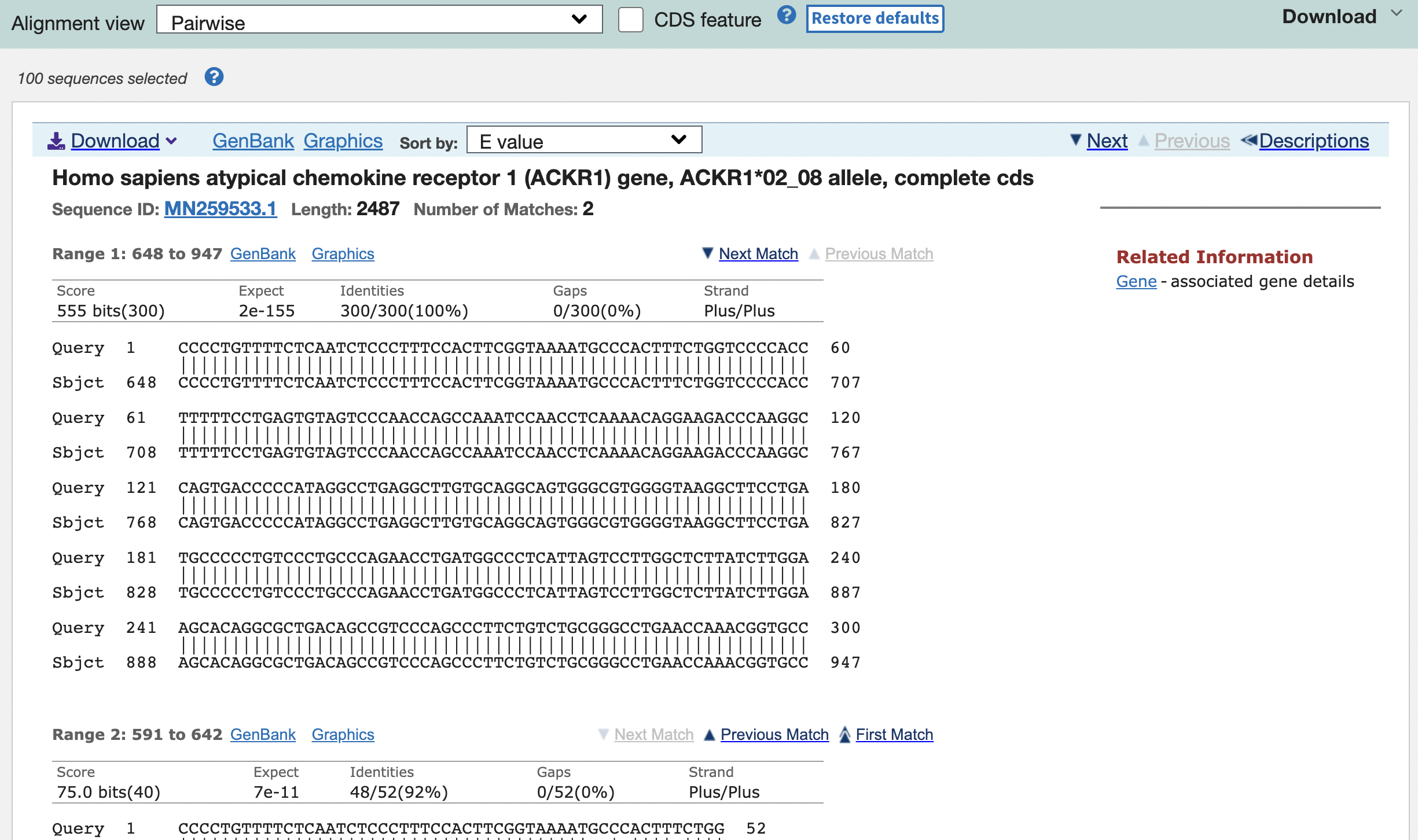
Congratulations! You have successfully run your first BLAST! From these results, the possibilities are limitless, so test out the algorithm and have a BLAST!





